Two new PyPI packages
Long time without posting… Let’s try to get back on track!
I have created two simple packages implementing useful probability distributions.
The LogUniform package provides the log-uniform and the modified log-uniform distributions, while the kumaraswamy package provides an implementation of the Kumaraswamy distribution.
Both packages implement similar APIs to the scipy.stats package.
LogUniform
The log-uniform distribution, often called the reciprocal distribution (Wikipedia) or, in some contexts, the Jeffreys prior, is commonly used as a prior for parameters which vary over several orders of magnitude.
Its probability density function (pdf) is given by
\[f( x | a,b ) = \frac{ 1 }{ x \, [ \ln( b ) - \ln( a ) ]} \quad \text{ for } a \le x \le b \text{ and } a > 0.\]The two parameters \(a\) and \(b\) correspond to the lower and upper bounds of the support.
Here is what it looks like, for \(a=1\), \(b=100\)

The modified log-uniform distribution is a modified version of the log-uniform distribution (!), which extends its support to include \(x=0\). The pdf is
\[f( x | x_0,b ) = \frac{ 1 }{ (x + x_0) \, \ln \left( \frac{b}{x_0} + 1 \right) } \quad \text{ for } 0 \le x \le b \text{ and } 0 < x_0 < b.\]with \(x_0\) (sometimes called the knee of the distribution) and \(b\) as parameters.
The support of this modified distribution goes from \(0\) to \(b\).
It looks like this, for \(x_0=1\) and \(b=100\)

Ok, now some code. To use the LogUniform implementation1, we would first install the package
$ pip install LogUniformand then use it from Python as
import loguniform
dist = loguniform.LogUniform(a=1, b=100)The dist object has methods much like those of a scipy.stats distribution:
dist.pdf(x) # Probability density function evaluated at x
dist.cdf(x) # Cumulative distribution function (cdf) evaluated at x
dist.sf(x) # Survival function (1 - `cdf`) evaluated at x
dist.ppf(p) # Percent point function (inverse of `cdf`) evaluated at pand a few extra properties:
>>> dist.mean
21.497576854210962
>>> dist.mode
1
>>> dist.std, dist.var
(24.96961794931886, 623.4818205349466)
>>> dist.skewness, dist.kurtosis
(1.4283873461330863, 1.0492789588735452)The same thing for the modified log-uniform:
dist = loguniform.ModifiedLogUniform(knee=1, b=100)>>> dist.pdf(0)
array(0.21667907)
>>> dist.cdf(dist.b)
1.0>>> dist.mean
20.667906533553168
>>> dist.mode
0
>>> dist.std, dist.var
(25.210415697969275, 635.5650596644156)
>>> dist.skewness, dist.kurtosis
(1.430935496096598, 1.0573553176553114)kumaraswamy
The Kumaraswamy distribution (see Wikipedia) is a little known family of continuous probability distributions defined on the interval [0,1]. It’s particular because of the similarities to the Beta distribution, and because (unlike the Beta) its pdf and cdf are easily evaluated.
The pdf is given by
\[f(x | a,b) = a \, b \, x^{a-1}{ (1-x^a)}^{b-1}, \quad \text{ for } 0 \le x \le 1 \text{ and } a>0, b>0,\]with \(a\) and \(b\) the two shape parameters. Depending on the values of these parameters, the distribution can have a range of different shapes:
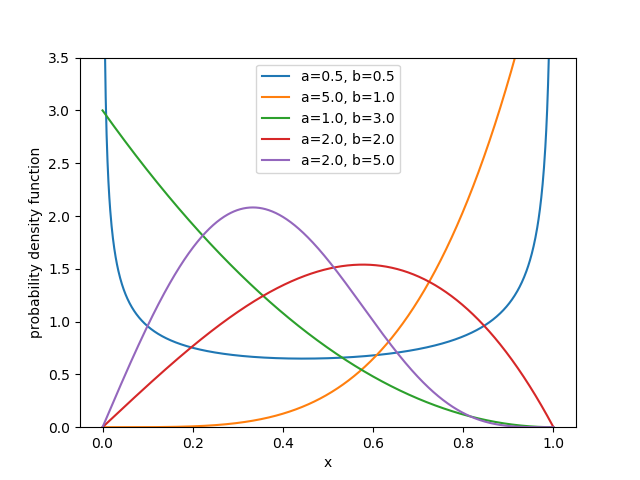
In order to use the kumaraswamy package, we first install it
$ pip install kumaraswamyand then use it from Python, much like before
import kumaraswamy
dist = kumaraswamy.kumaraswamy(a=.5, b=.5)with the same methods and properties now available in dist.
To finish, one extra method shared by the three distributions provides random samples:
>>> dist = loguniform.LogUniform(a=1, b=100)
>>> dist.rvs(3)
array([99.88014766, 28.10705983, 34.18542689])
>>> dist = loguniform.ModifiedLogUniform(knee=1, b=100)
>>> dist.rvs(100)
array([1.30284699e+01, 3.26860325e+01, 1.10717848e+00, 8.66021941e+01,
3.66928668e+01, 6.40408882e+00, 1.50455730e+01, 3.90749222e+01,
...
>>> dist = kumaraswamy.kumaraswamy(a=.5, b=.5)
>>> dist.rvs()
0.4416781513613892wrap up
That’s it, two simple packages implementing three probability distributions for some of your statistical needs.
Let me know in the comments if this is helpful, wrong or super-duper cool.
-
Note that the log-uniform distribution is already implemented in
scipy.stats.reciprocal, but not the modified distribution. ↩